Experiencing science
in our everyday lives
Intro | Director’s Message | Reader's Guide

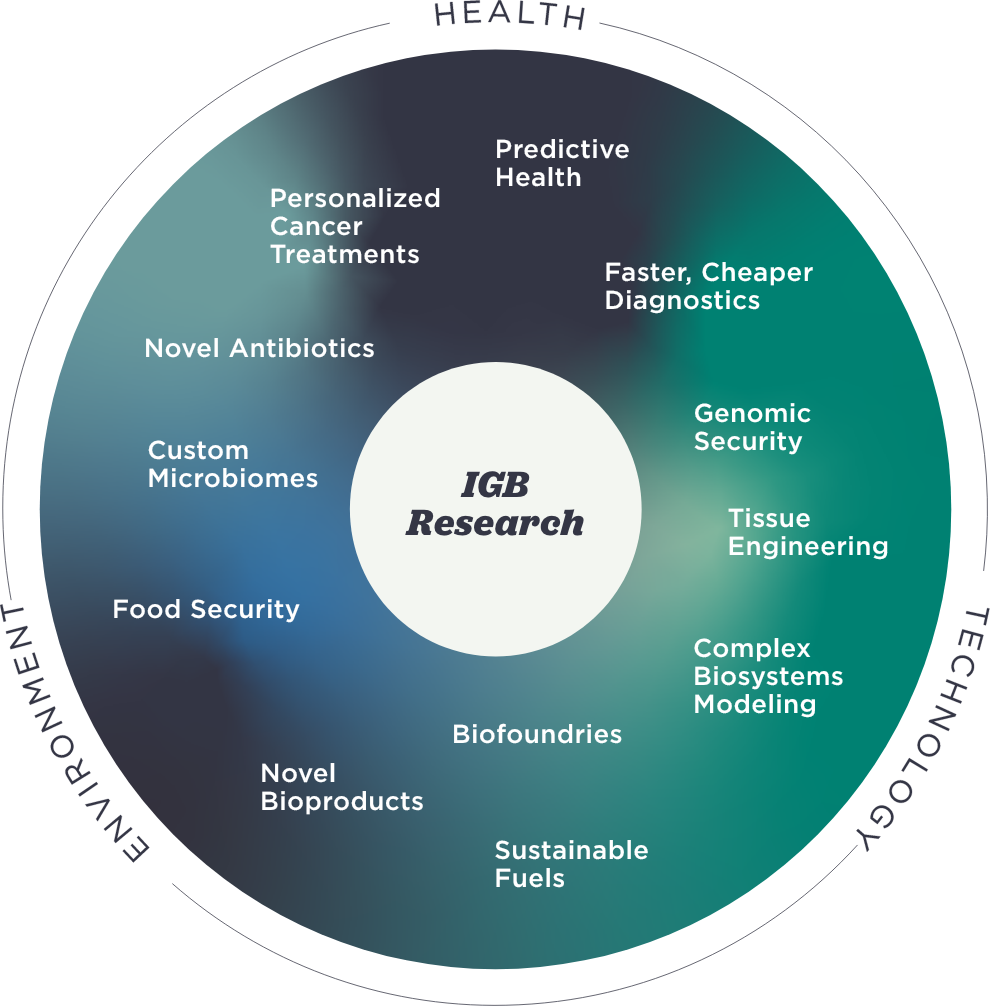
IGB
at a Glance
2019 Highlights

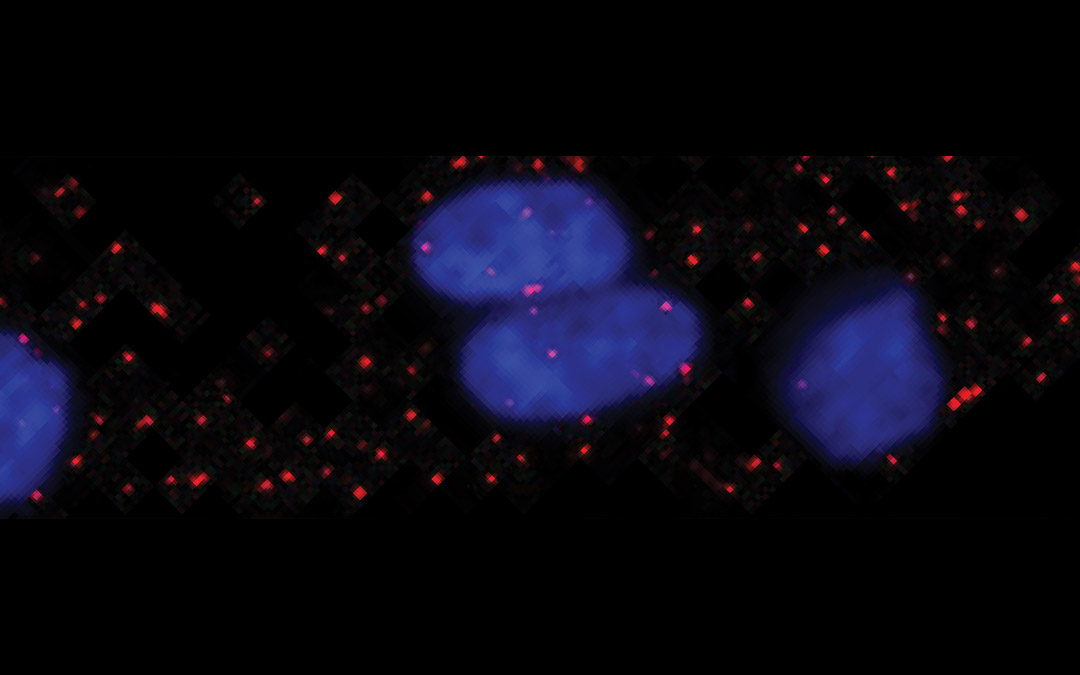
Can Genomics
help me be a
Smarter Shopper?

Can Genomics
Help Me Understand my
Family Dynamics?
Can Genomics
Help Me Optimize My
Workout Routine?

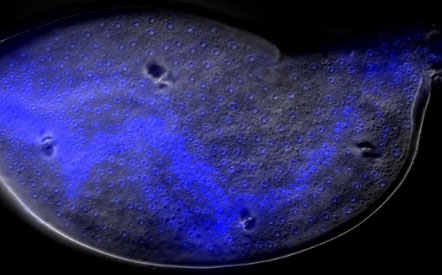
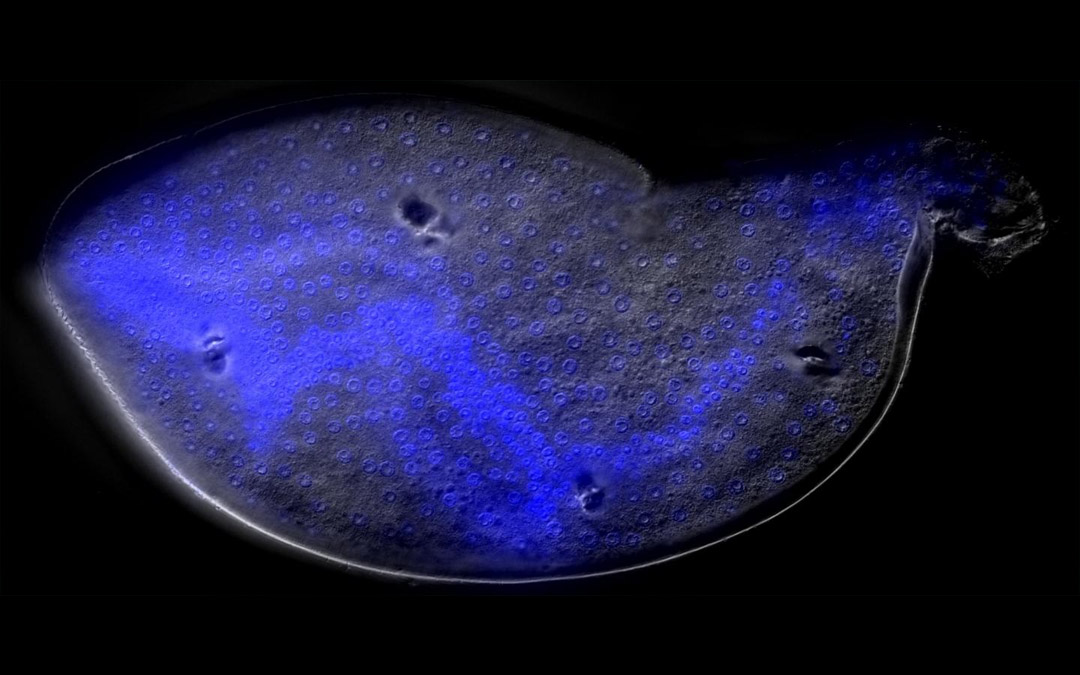
Can Genomics
help me to appreciate and protect the
Natural World?

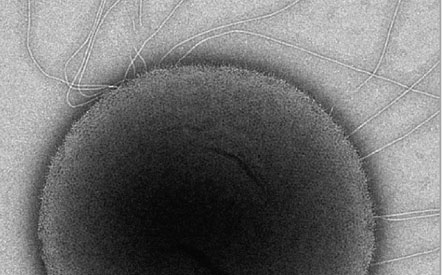
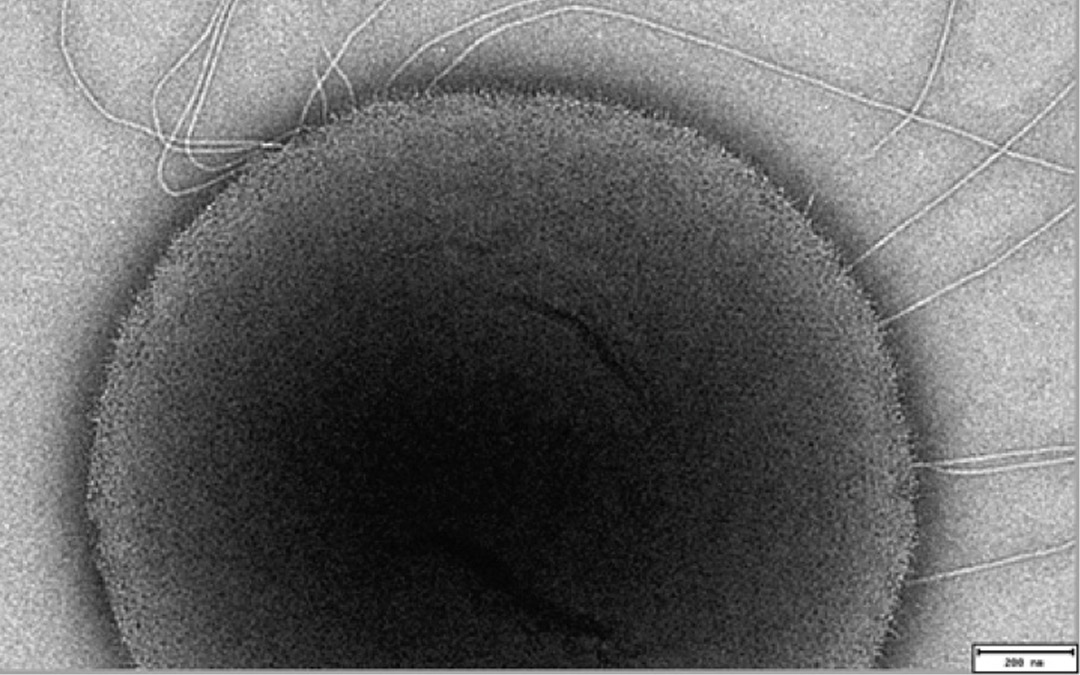
Can Genomics
help me to
Feel Better?
By the
Numbers
Opportunities, growth, and honors