
Art of Science 5.0
April 10, 2015 - April 10, 2015
CURATED BY
Kathryn Faith
WHEN
April 14 - April 14
WHERE

View Gallery
1
/
10
1 / x

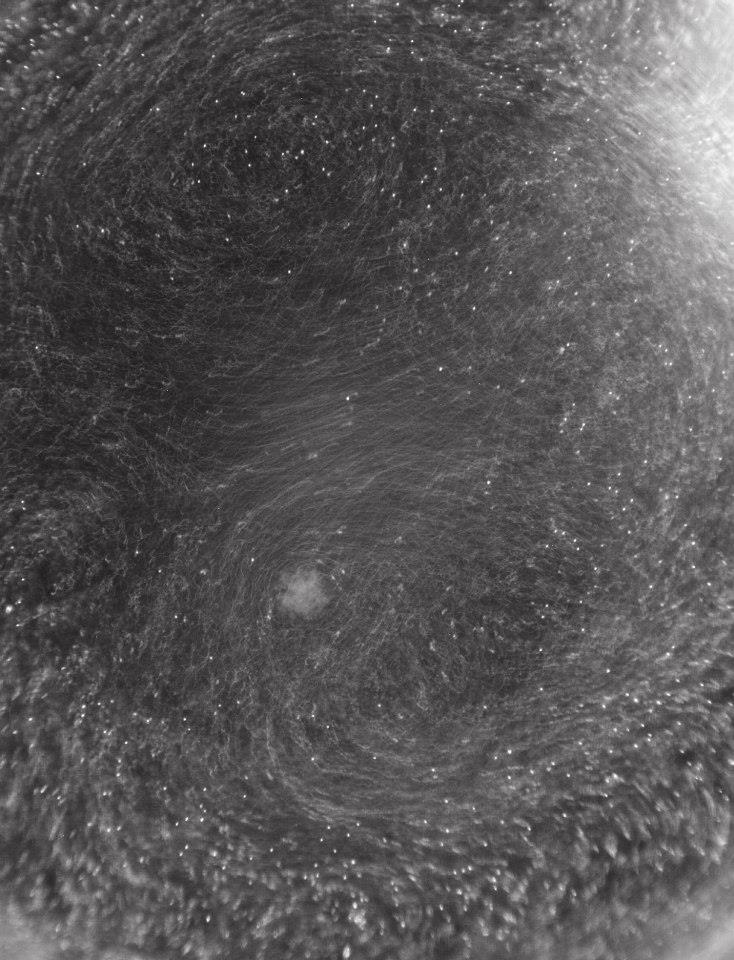
Nebula
This sparkling cloud is actually an aggregate of oil and fine sediment that occurs when water, fine sediment, and an oil film is mixed. The bright white circles are droplets of oil that fluoresce under ultraviolet light. The darker material surrounding the droplets is a complex group of fine sediment grains that reflect visible light. Examining how this type of aggregate forms can help researchers understand how spilled oil lingers in the environment and affects wildlife and human health.

Tapestry of Youth
An intermediate stage in muscle development is the formation of myotubes, long thin structures that will grow into muscle fibers. The patterns seen in this image are created by myotubes grown in a gel matrix. Observing how myotubes develop in laboratory conditions aids in the understanding of how skeletal muscle tissue develops. This knowledge can help us to better understand how the environment around cells guides development, and how to treat diseases that affect the muscles.
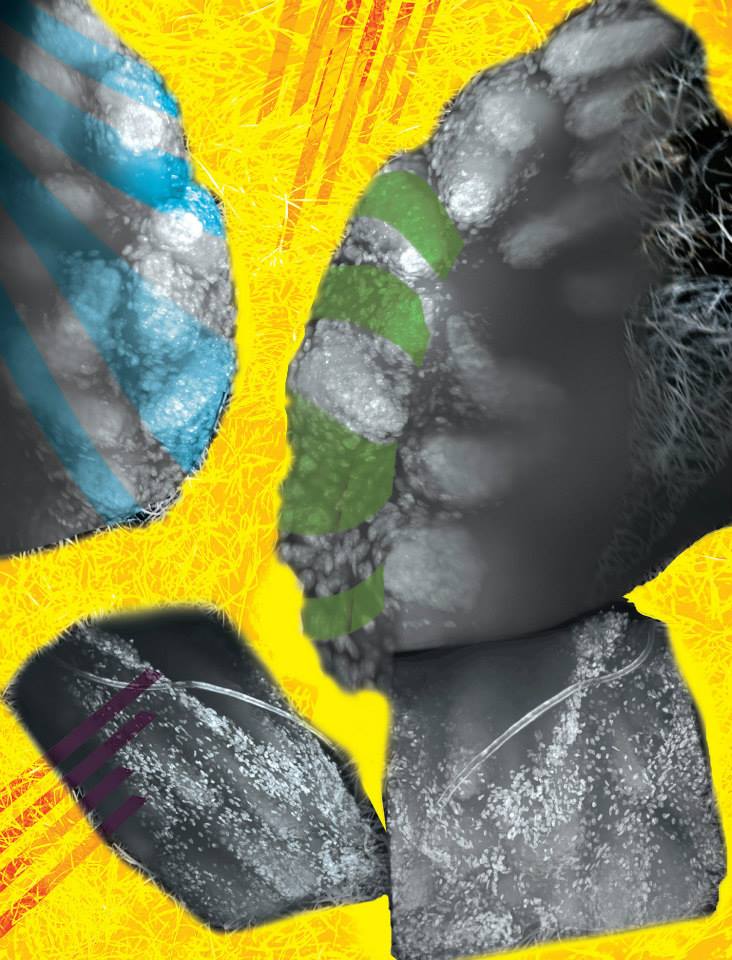
Fighting for Breath
This sparkling cloud is actually an aggregate of oil and fine sediment that occurs when water, fine sediment, and an oil film is mixed. The bright white circles are droplets of oil that fluoresce under ultraviolet light. The darker material surrounding the droplets is a complex group of fine sediment grains that reflect visible light. Examining how this type of aggregate forms can help researchers understand how spilled oil lingers in the environment and affects wildlife and human health.

Step by Step
Calcite, a component of sedimentary rock such as limestone, forms in natural environments such as Yellowstone National Park. This image shows the surface of a growing crystal of calcite; the high-power microscope used to produce the image allows the visualization of individual molecular layers, revealing new details of the calcite crystal formation process.

Cell Wars
The bright, contrasting colors in this image highlight the battle lines drawn by single-celled microorganisms, engaged in chemical warfare at a scale invisible to the human eye. Related species continually fight each other for access to food and safe environments to grow and multiply. By labeling two competing types of bacteria with two different dyes, researchers were able to track the progress of their conflict.
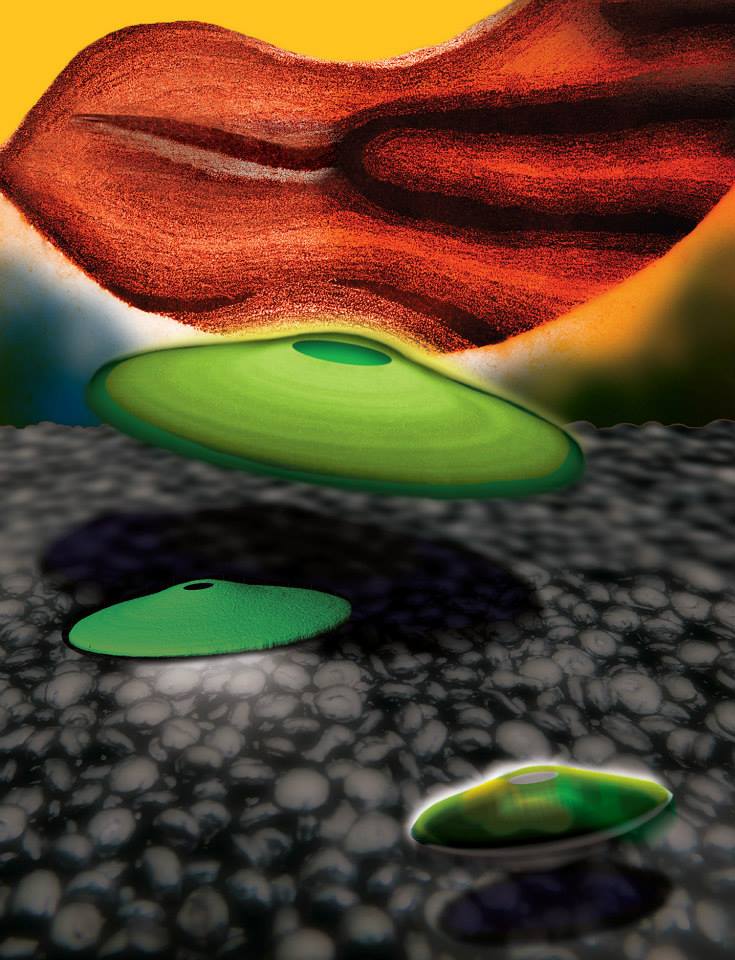
Looking Through Walls
The prostate is a small and complicated structure that plays an important role in the male reproductive system. This image shows several angles of a mouse prostate gland, prepared and dyed so that the cellular structure of the whole organ can be seen and recorded. The 3D images created by this work will contribute to understanding of tissue development.

Home Fires
This image shows human mammary gland cells expressing a genetically-encoded fluorescent dye. The dye is designed to respond to the metabolic state of structures inside the cell; in this case, it is labelling mitochondria, the energy-generating structures used by plant, animal, and fungal cells. Images like this one show how abundant mitochondria are inside the cell, and the complexity of their activity. This research offers novel insight into basic cellular functions that are altered in cancer.
Working as One
This image offers experimental evidence for a theoretical model of how E. coli cells cooperate to use resources. The model predicted that oxygen becomes less available to cells inside the colony as it grows, and sugar is less available to cells at the top; cells near the base of the colony are expected to break down glucose into acetate, which would then be consumed by cells near the top. A dye used to label living colonies shows that cells near the surface do upregulate genes associated with acetate consumption, just as the model predicted.

Identities
This sparkling cloud is actually an aggregate of oil and fine sediment that occurs when water, fine sediment, and an oil film is mixed. The bright white circles are droplets of oil that fluoresce under ultraviolet light. The darker material surrounding the droplets is a complex group of fine sediment grains that reflect visible light. Examining how this type of aggregate forms can help researchers understand how spilled oil lingers in the environment and affects wildlife and human health.

Cross My Heart
Every day, the human heart pumps approximately 7,570 liters of blood and beats approximately 100,000 times. The pump rate responds to internal and external stimuli such as exercise, medications, diet, age and heart disease. This image of a mouse heart was created as part of a study to investigate and better understand changes in whole organ structure, chamber dimension and wall thickness of the mouse heart during the development of heart disease.

Battle Testing
This sparkling cloud is actually an aggregate of oil and fine sediment that occurs when water, fine sediment, and an oil film is mixed. The bright white circles are droplets of oil that fluoresce under ultraviolet light. The darker material surrounding the droplets is a complex group of fine sediment grains that reflect visible light. Examining how this type of aggregate forms can help researchers understand how spilled oil lingers in the environment and affects wildlife and human health.

The Key to Skeletons
This image was produced as part of a project that focuses on a crucial, yet often overlooked, aspect of the evolutionary transition from reptiles to mammals. Over evolutionary time, bones in the reptilian jaw joint have migrated into the middle ear of mammals and become co-opted for hearing. To examine the development of bone structure, the mouse fetus shown here was treated to make the tissue transparent, and dyed to label the cartilage and bone.

Mathematical Forestation
This is an image of tree shapes that emerge from competition for sunlight in a computational evolution experiment. To prepare for the possibility that other trees may overshadow them, trees are driven to become taller despite the energetic cost of growth. Computational open-ended evolution strategies allow researchers to create simulations that lead to new discoveries.

Lifelines
The iBioFAB is an integrated robotic system designed for large-scale DNA editing and cell screening. This image is based on the output of iBioFAB as it continuously monitored the cell density of a yeast strain. iBioFAB applies automation to quickly perform experiments that would otherwise take months of work. This technology can potentially accelerate the development of new drugs, better biofuel-producing microbial strains, and other biotechnological innovations.

In a Single Breath
To create this image, researchers used specialized software to track the movement of fluorescent beads and calculate a mean speed of movement. This is a way to quantify the ability of cilia, tendril-like cellular structures that act like paddles or appendages, to move mucus inside the human lung. In cystic fibrosis, these cilia are impeded by thick mucus and cannot move. This experiment is a way to study potential treatments to restore normal lung function in individuals with cystic fibrosis.

Share and Share Alike
How do new genes arise? Microbes live in diverse and extreme environments —from deep-sea hydrothermal vents, to salt marshes, to the human gut. To survive, microbes obtain genes from other organisms, essentially stealing evolutionary solutions for living in challenging environments. Each dot in this image represents a single gene from one of several strains of bacteria. The dots’ colors and positions highlight similarities and differences in the ways that the genetic code, the correspondence between DNA sequence and protein structure, is used in each gene. This study has given researchers new perspective on how microbes share novel and disease-associated genes.

Defense Mechanisms
This image shows gene activity in the mouse brain in response to encountering an intruder for the first time. House mice, stickleback fish, and honey bees are very different animals, but when researchers closely examine the molecular responses of each species when exposed to danger, those responses are remarkably similar. All three species increase the activity of genes that regulate hormones and neurotransmitters affecting their behavior. This suggests that organisms share a common toolkit of gene activity that guides their behavior.

Animal, Vegetable, Mineral
The lacy patterns in this image are part of a paper-thin section of travertine limestone collected from Mammoth Hot Springs in Yellowstone National Park. The travertine is composed of filaments of bacteria around which tiny crystals of the mineral aragonite have formed. In order to examine these delicate samples, the minerals are embedded in epoxy and polished down to a thin section. The research that produced this image examines the ways in which water, minerals, and microbes interact with one another in a hot spring environment.
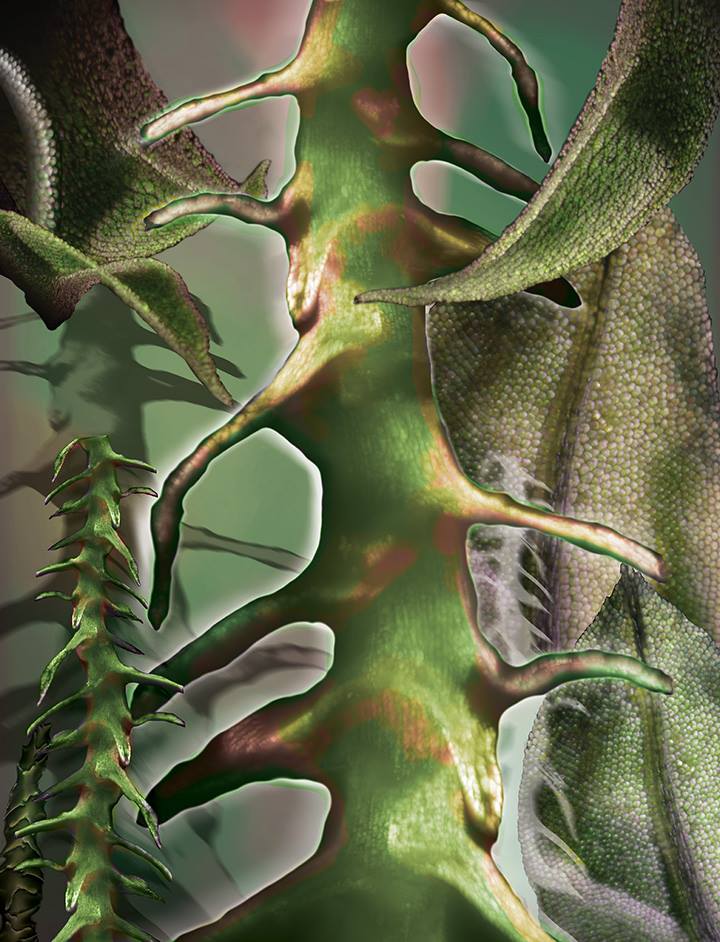
Light and Shade
This collage of images shows microscope structures within moss collected in Urbana, Illinois, as part of a study to examine light movement through moss. Mosses are small, non-flowering plants. They are typically found in shaded, damp places and on the sides of trees. Like all plants, they rely on photosynthesis to make energy from sunlight. However, unlike leaves of most other plants, moss leaves typically are a single layer of cells. This anatomical difference greatly alters the way light moves through the leaf and canopy.

It’s the Little Things
These fancifully shaped orange and yellow modes represent individual pollen grains from a water-willow plant Justicia asclepiadea and white spruce Picea glauca respectively. A team of middle school girls participating in the IGB Pollen Power! summer camp created images of the pollen grain, which were then transformed into a 3D computer model and printed in plastic at the University of Illinois College of Business MakerLab. This is one of several hands-on activities through which camp participants learn about plant biology. The grant that provides support for the camp also funds research on development of ozone-resistance in corn.
Nebula
Scientist Collaborator
David Waterman
Marcelo H. Garcia Laboratory
Ven Te Chow Hydrosystems Laboratory
Instrument
Zeiss Axioobserver with the Apotome
Funding Agency
Funded by the U.S. Environmental Protection Agency
Original Imaging



Special Thanks
Tapestry of Youth
Scientist Collaborator
Max Rich
Hyunjoon Kong Laboratory
Instrument
Multiphoton Confocal Microscope Zeiss 710 with Mai Tai eHP Ti:Sapphire Laser
Funding Agency
Funded by an NSF Science and Technology Center
Original Imaging



Special Thanks
Fighting for Breath
Scientist Collaborator
Katrina Diaz
Martin Burke Laboratory
Instrument
Zeiss Axiozoom V16 Scope
Funding Agency
Funded by the HHMI and NIH
Original Imaging



Special Thanks
Step by Step
Scientist Collaborator
Xudong Guan
Core Facilities & Microscopy Suite
Instrument
Cypher ES Atomic Force Microscope
Funding Agency
Funded by the Carl R. Woese Institute for Genomic Biology
Original Imaging



Special Thanks
Cell Wars
Scientist Collaborator
Venhar Celik
Ting Lu Laboratory
Instrument
Zeiss Stereolumar v12
Funding Agency
Funded by the University of Illinois
Original Imaging



Special Thanks
Looking Through Walls
Scientist Collaborator
Xiaochen Lu
Lisa Stubbs Laboratory
Instrument
Zeiss Lightsheet Z.1. Imaris 3D Visualization Package
Funding Agency
Funded by the NIH
Original Imaging



Special Thanks
Home Fires
Scientist Collaborator
Matthew Leslie
Rex Gaskins and Paul Kenis Laboratories
Instrument
Zeiss Axioobserver with the Apotome
Funding Agency
Funded by the NIH
Original Imaging



Special Thanks
Identities
Scientist Collaborator
Jennifer Mitchell
Martha Gillette Laboratory
Instrument
SR-SIM - Four Laser Structured Illumination System
Funding Agency
Funded by the NSF
Original Imaging



Special Thanks
Cross My Heart
Scientist Collaborator
Sakthivel Sadayappan
Sakthivel Sadayappan Laboratory at Loyola University Chicago
Instrument
Multiphoton Confocal Microscope Zeiss 710 with Mai Tai eHP Ti:Sapphire Laser
Funding Agency
Funded by the NIH
Original Imaging



Special Thanks
Battle Testing
Scientist Collaborator
Sara Pedron and Bhushan Mahadik
Brendan Harley Laboratory
Instrument
Zeiss Lightsheet Z.1; Imaris 3D Visualization Package
Funding Agency
Funded by the NSF, NIH, and Mayo Clinic-University of Illinois Alliance
Original Imaging



Special Thanks
The Key to Skeletons
Scientist Collaborator
Daniel Urban
Karen Sears Laboratory
Instrument
Zeiss Axiozoom V16 Scope; Zen Pro 2012 Software
Funding Agency
Funded by the NSF
Original Imaging



Special Thanks
Mathematical Forestation
Scientist Collaborator
Nicholas Guttenberg
Nigel Goldenfeld Laboratory
Instrument
Blender, Open Source 3D Rendering Package
Funding Agency
Funded by the NSF
Original Imaging



Special Thanks
Lifelines
Scientist Collaborator
Ran Chao
Huimin Zhao Laboratory
Instrument
Illinois Biological Foundry for Advanced Biomanufacturing (iBioFAB)
Funding Agency
Funded by the Roy J. Carver Charitable Trust and the Carl R. Woese Institute for Genomic Biology
Original Imaging



Special Thanks
In a Single Breath
Scientist Collaborator
Katrina Diaz and Alex Cioffi
Martin Burke Laboratory
http://www.scs.illinois.edu/burke/
Instrument
Zeiss Axioobserver with the Apotome; Imaris 3D Visualization Package
Funding Agency
Funded by the NIH and HHMI
Original Imaging



Special Thanks
Share and Share Alike
Scientist Collaborator
Katherine Karberg and Gary Olsen
Gary Olsen Laboratory
Instrument
Codon Usage Utilties Software; POV-Ray Visualization Software
Funding Agency
Funded by the NASA and the DOE
Original Imaging



Special Thanks
Defense Mechanisms
Scientist Collaborator
Derek Caetano-Anollés and Jacqueline Brinkman
Lisa Stubbs Laboratory
Instrument
NanoZoomer Slider Scanner
Funding Agency
Funded by the Simons Foundation
Original Imaging



Special Thanks
Animal, Vegetable, Mineral?
Scientist Collaborator
Laura DeMott
Bruce Fouke Laboratory
Instrument
NanoZoomer Slider Scanner
Funding Agency
Funded by NASA
Original Imaging



Special Thanks
Light and Shade
Scientist Collaborator
Aleel K. Grennan
Donald R. Ort Laboratory
Instrument
Zeiss Lightsheet Z.1; Imaris 3D Visualization Package
Funding Agency
Funded by DOE
Original Imaging



Special Thanks
It's the Little Things
Scientist Collaborator
IGB Pollen Power Camp for Middle School Girls
Andrew Leakey, Glenn Fried and Mayandi Sivaguru
Core Facilities & Microscopy Suite
Instrument
Multiphoton Confocal Microscope Zeiss 710 with Mai Tai eHP Ti:Sapphire Laser; Imaris 3D Visualization Package
Funding Agency
Funded by the NSF and the Carl R. Woese Institute for Genomic Biology
Original Imaging



Special Thanks
Special Thanks
Champaign businessman Doug Nelson, President of BodyWork Associates, first proposed the idea that became Art of Science, and his continued efforts to support the exhibit made its realization possible. The IGB is also grateful to James Barham of Barham Benefit Group and [co][lab] founder Matt Cho for hosting the annual exhibit.


