Carl R. Woese Institute for Genomic Biology


The image is showing a two-dimensional (2D) DNA origami self-assembled of synthetic DNA oligonucleotides with programmed sequences. This is a molecular pegboard/platform (80nm*80nm) that can arrange external elements with nanometer scale accuracy for different applications. The sample was scanned using Cypher AFM from Asylum Research at the Carl R. Woese Institute for Genomic Biology Core Facility.
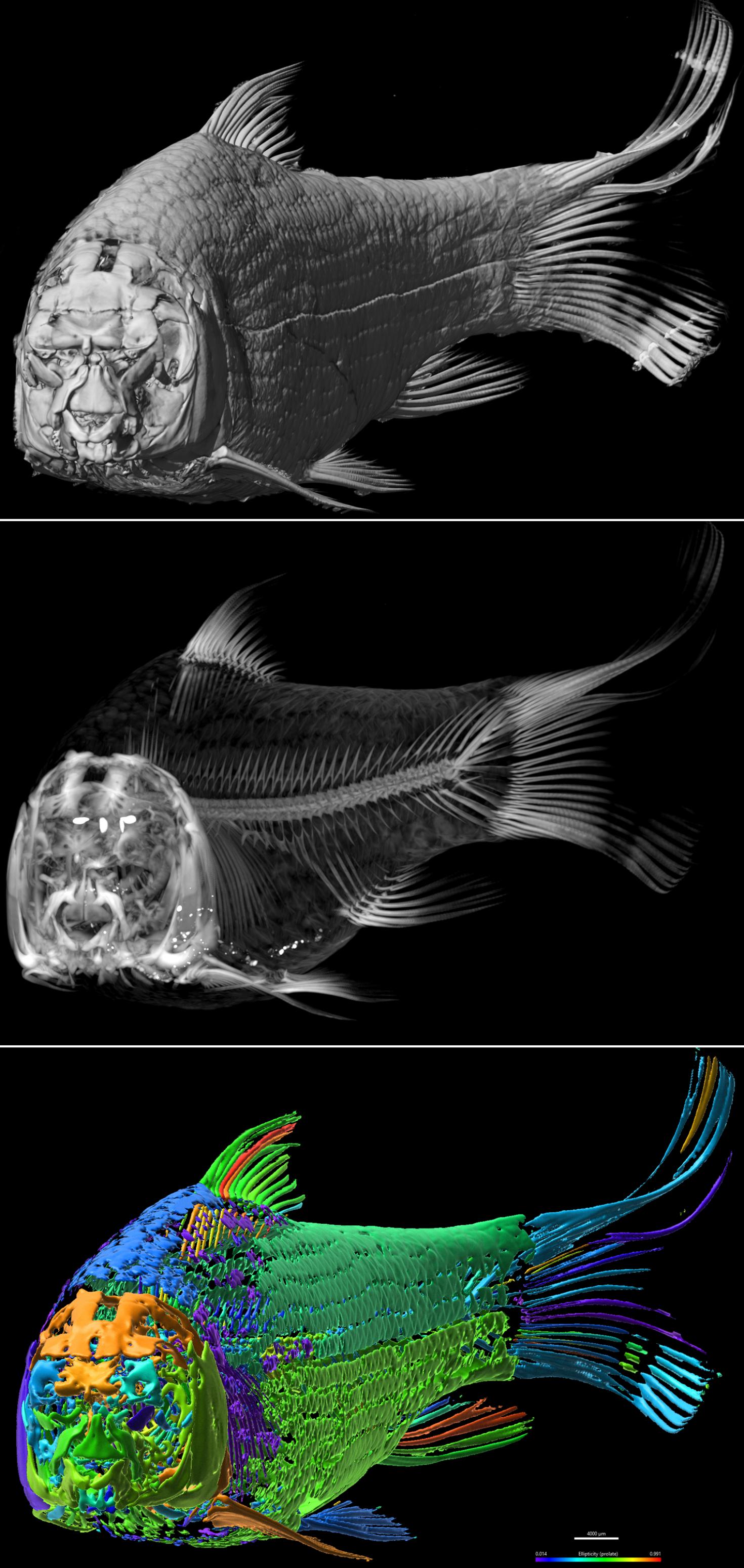

The image is showing a Shorthead Redhorse, Moxostoma macrolepidotum. The species is commonly found in large creeks and rivers across most of Illinois. It is a member of the "sucker" family, Catostomidae, whose name originates from the ability to use their mouths to suck up small food items usually found on creek and river bottoms. It can reach a maximum size up to 2 1/2 feet in length. The image sequence from top to bottom shows 3D volume rendering at different opacities/modules to show external surface (top), internal skeleton (middle) and psuedocolored Isosurface objects with range of ellipticity (prolate, color coded) from Xray CT data. The sample was scanned using the NSI X5000 High Resolution MicroCT and rendered in 3D using Imaris 3D visualization program at the Carl R. Woese Institute for Genomic Biology Core Facility.
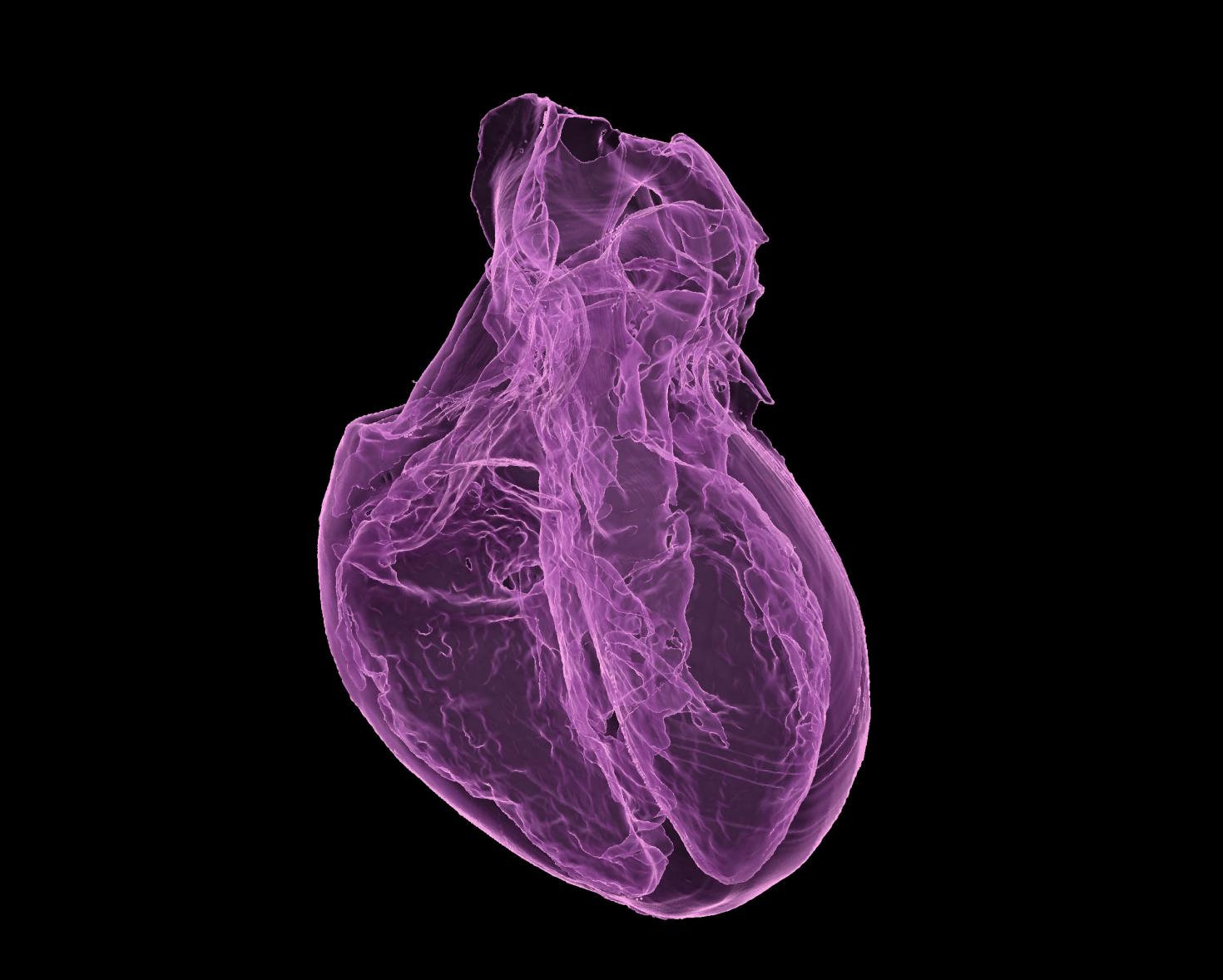
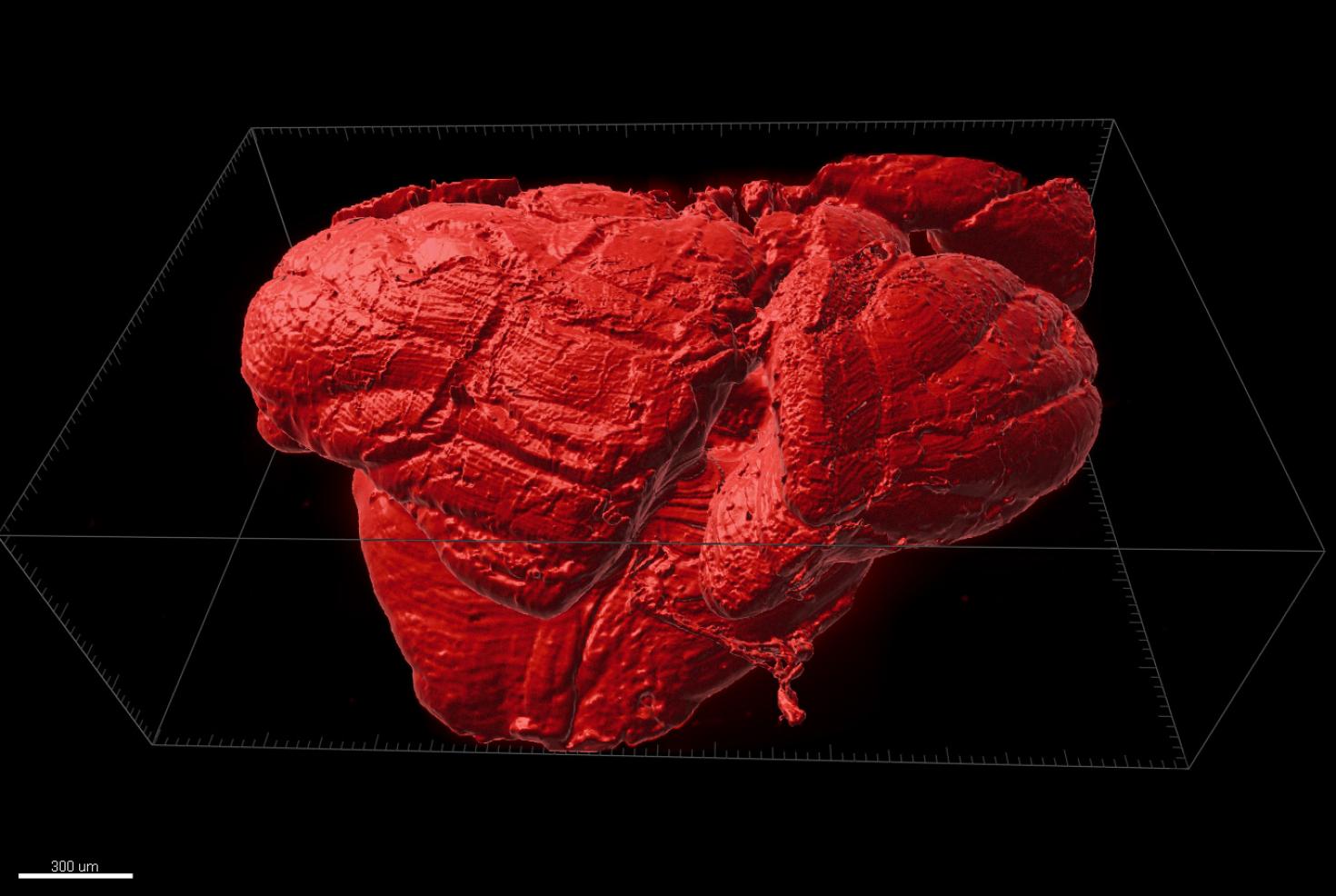
The image is showing a 3D printed non-drying hydrogel (polyacrylamide, water and glycerol) heart model. This is a prototype measuring approximately 3.5x2.5x2.5 cm to check the integrity of right and left ventricles of the heart. Eventually, a larger model will be printed and be used as suture practice platform for surgeons. The 3D rendered image pseudocolored purple using Isosurface module from Xray CT data having 24-micron voxel size. The sample was scanned using the X5000 NSI High Resolution MicroCT and rendered in 3D using Imaris 3D visualization program at the Carl R. Woese Institute for Genomic Biology Core Facility. Image provided by Joanne Vanessa Hwang from Hyunjoon Kong Group .
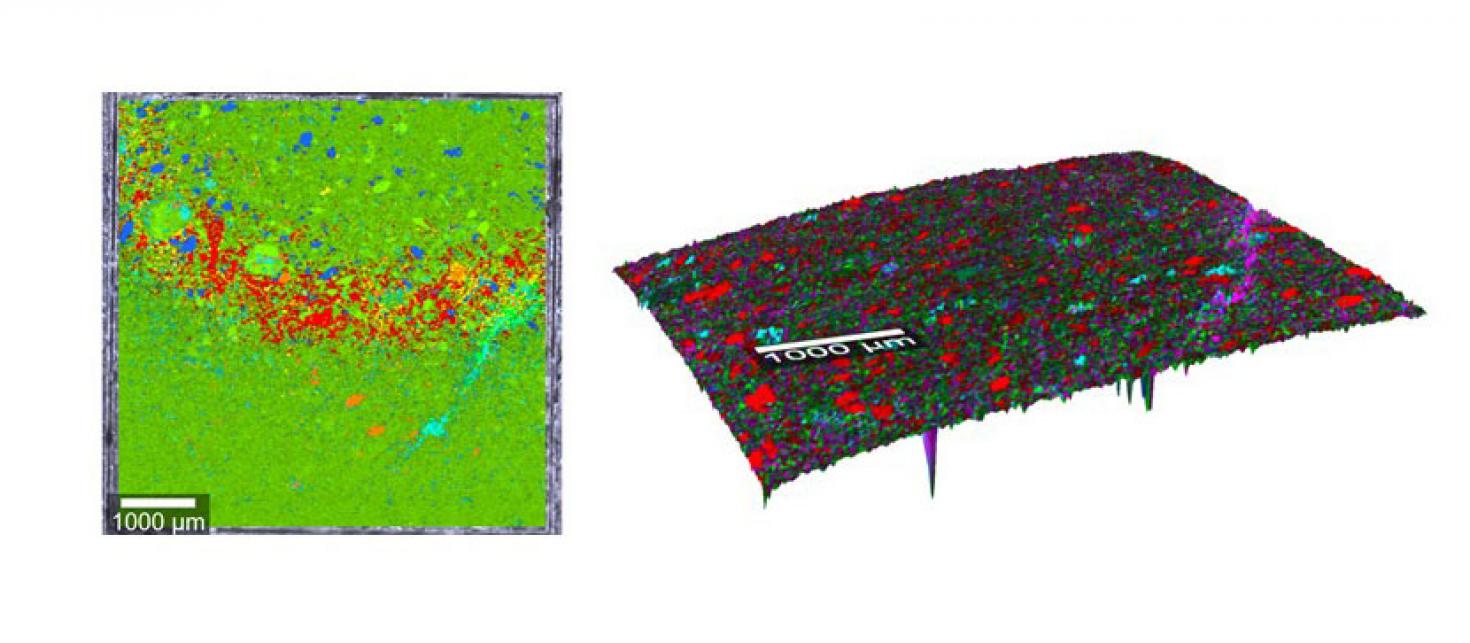

Raman composite image overlaid on surface contour showing different minerals of a polished construction rock. The sample was scanned using the WiTech AFM-Raman spectrometer using TruSurface module at the Carl R. Woese Institute for Genomic Biology Core Facility. Image provided by Krishna Chaitanya Polavaram from the Garg Group. The left image is without Trusurface module and the right is with Trusurface Raman, where the entire contour was in focus.
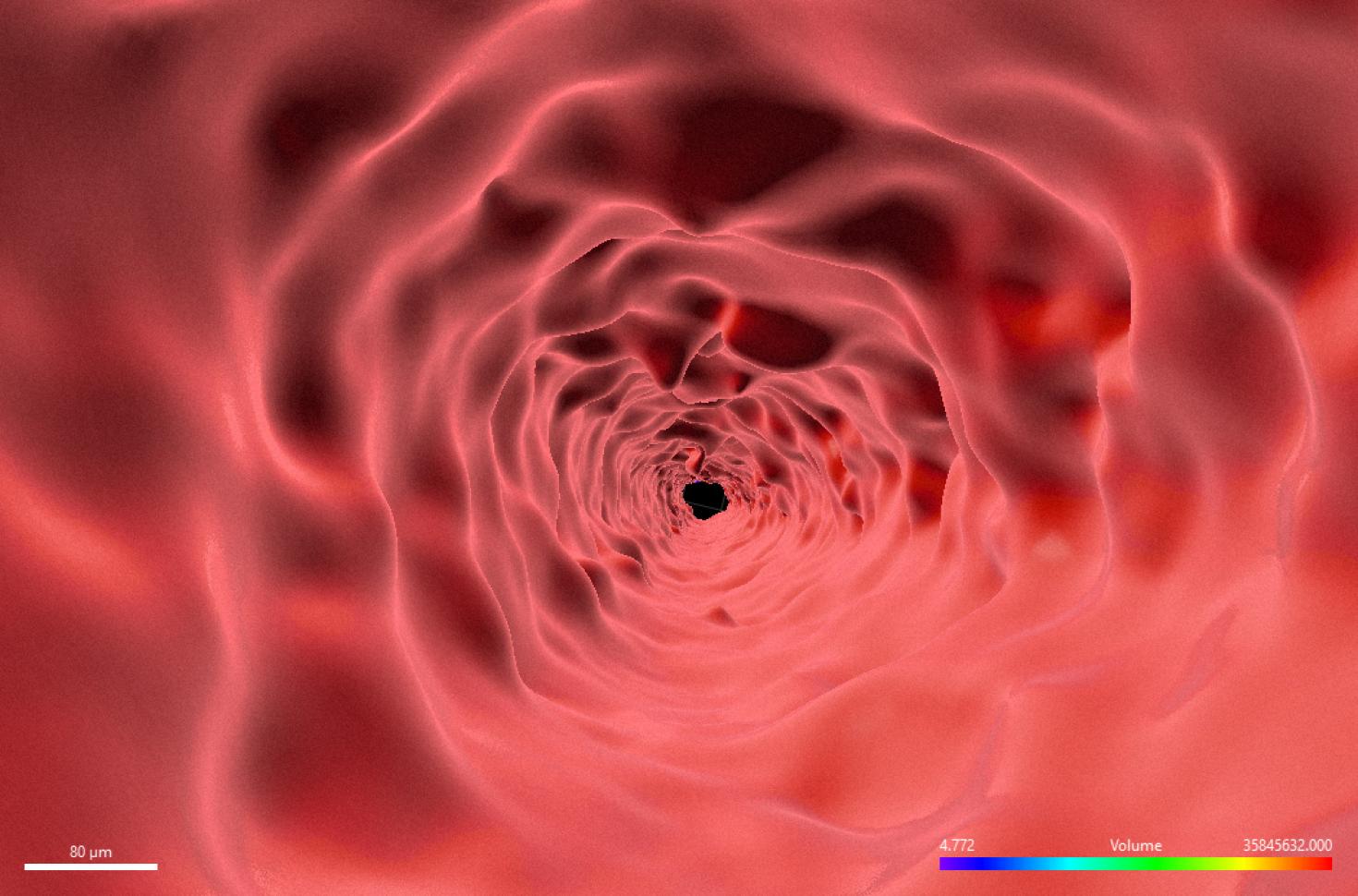
This image is of an inside contour surface of a 1000-micron aluminum tube produced by direct metal laser sintering (DMLS). Reconstructed surface contours like this created in Imaris from X5000 Xray CT data, are used to compare the geometrical parameters of the printed object to the design. The sample was scanned using the X5000 NSI High Resolution MicroCT and rendered in 3D using Imaris at the Carl R. Woese Institute for Genomic Biology Core Facility. Image provided by Kevin Uvodich from the Nenad Miljkovic Group.

Half-Mouse Brain: This tile scan of a whole brain of a cuprizone-fed mouse was obtained with an Ultra Microscope (a 2nd generation lightsheet microscope by Miltenyi Biotec) after CLARITY tissue clearing and subsequent staining with anti-proteolipid protein (PLP). The image was processed with the Imaris 3D Software System: UltraMicroscope II - Miltenyi Biotec. Sample provided by Allison Yukiko Louie, Dr. Steelman's Laboratory, Department of Animal Sciences and imaged/proceed by Dr. Kingsley Boateng, Carl R. Woese Institute of Genomic Biology, UIUC.


Fossil Insect in Amber: This is a fossil earwig (order Dermaptera) preserved in amber from the Dominican Republic. The amber is Early Miocene in age, between 16 and 18 million years old. The preservation of fossil insects in Dominican amber is exquisite and often includes such fine morphological features as wing venation, delicate hairs/setae, and individual lenses, or ommatidia, of the compound eyes. In this specimen the eyes, antennae, and folded wings are clearly visible, along with the forceps, or pincers, at the end of the abdomen that are characteristic of earwigs. It is also possible to count the number of plates on the underside of the abdomen which indicates that this specimen is an adult female.
System: NSI X5000 High Resolution MicroCT System

View Gallery
1
/
10
